Introduction
Data Preparation
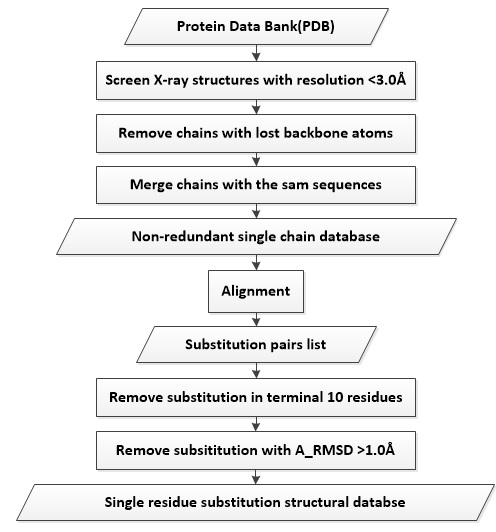
To construct the database of structure pairs of single residue substitution, we downloaded all the protein X-ray structures with resolution < 3.0 Å from Protein Data Bank (PDB) of the edition 17th. The protein structures were split into single chains (subunits), and renamed by the PDB code and the chain ID. For example 1a2cA indicates the chain A of PDB 1a2c. In addition, protein subunits with backbone atom defects were removed, as the absence of main chain atoms will result in uncertain local structures. Besides, a number of proteins recur in the PDB database, such as the protein determined in different laboratory or the protein subunits exist in several complex. Protein subunits with the same sequence and RMSD < 1.0 Å were ranked by their resolutions. The one with the best resolution was selected to be representative. Thus, we collected a non-redundant single chain structure database with 45797 protein subunits.
Then we computed the residue substitution pairs with ProMut, resulting 11130 pairs. To reduce the influence by the thermal oscillations or disorder, residue substitutions occur in terminal residues (<10) were removed. The structure pairs with A_RMSD > 1.0 Å were also removed, because a large A_RMSD means that the structure pairs are not well superimposed, which may result in to much noise in M_RMSD. Those structure pairs with A_RMSD > 1.0 Å were collected in a subset for analyzing mutation related topology changes in our following work. Finally, we got 7419 structure pairs of protein residue substitution.
Query
The web server of the database was built the using PHP language. All the data was organized by MySQL database.
In the current release this resource can be browse by:
- Types of amino acid substitutions.
- Solvent accessibility of substituted residues.
- Backbone hydrogen bonds of substituted residues.
- Secondary structures of substituted residues.
- Ramachandran possibility of substituted residues.
- Side-chain packing compatability of substituted residues.
- PDB ID.
The search results are always returned in the form of a hit list represented by the site identifier, protein name and family, the list of all associated PDB entries. Clicking on the site identifier takes the user to the individual entry page.