Introduction
Antibody sequence numbering and complementarity determining region (CDR) delimitation have wide applications in antibody engineering. They are generally obtained by mapping query sequences to the retrospective patterns. However, due to the enormous diversity of antibody sequences, novel patterns are often generated in antibody affinity maturation. They may not be recognized by the traditional methods. Antibody Region-Specific Alignment (AbRSA) integrates the biological insight of antibody region-specific feature with dynamic programming to improve the robustness of antibody numbering. Benchmarks show AbRSA is a powerful method in numbering unusual antibodies and distinguishing between antibody and non-antibody sequences.
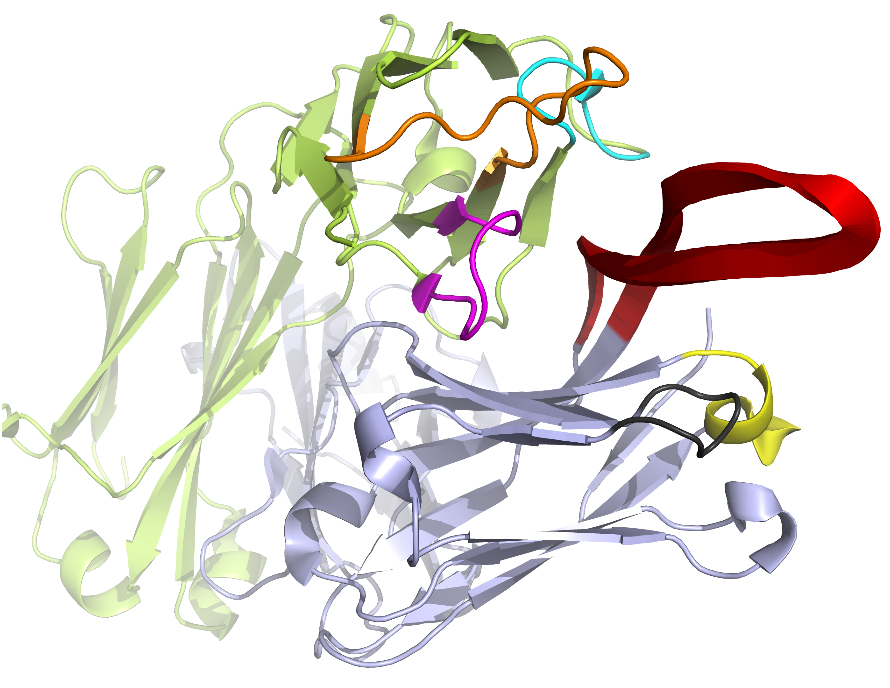
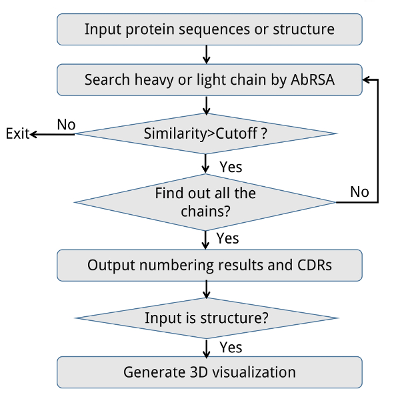
The pipeline of AbRSA web service is shown in the following Figure. The input could be either the protein sequence or structure. Multiple protein sequences are supported if the sequences are in FASTA format. The program judges whether it is a heavy chain, light chain or neither by comparing the sequence identities with consensus sequences. After all the possible heavy or light chains are found out, the program will output the numbering results and the location of FRs and CDRs in the sequences. If the input is a protein structure (PDB format), the web page will generate its interactive 3D visualization powered by 3Dmol JavaScript library (Rego and Koes, 2015). CDRs will be highlighted in colors. The 3D view can be rotated, translated, and re-sized by dragging, scrolling the mouse. We believe this feature could help to understand where and how antibody binding with antigen.
AbRSA web service is free for users, and, therefore, on an "as is" basis, without warranty of any kind.
Please Cite:
AbRSA: A robust tool for antibody numbering (https://doi.org/10.1002/pro.3633). 2019, Protein Science